JBiclustGE API
This section presents some examples of how to use JBiclustGE API, for helping the developers to use/extend the functionalities incorporated in this API.
For a detailed information of the classes implemented in this API, see the documentation.
Setup
JBiclustGE depends of several R packages to work properly, thus in the first time that JBiclustGE is executed needs to be always configured concerning to some operational aspects such as: R settings (mainly the folder to install the R packages), maximum allowed p-value, and other optional settings. There is an special class (JBiclustGESetupManager.class) that is responsible to configure, download the required algorithms/R packages, and build the file (jbclustge.properties) that stores the settings of the application.
Important methods of class JBiclustGESetupManager:
-
Check if JbiclustGE Environment its configured:
boolean isconfigured= JBiclustGESetupManager.isJbiclustGEConfigured(); -
Setup the required algorithms in JBiclustGE:
JBiclustGESetupManager.setupJBiclustGEMethodsEnvironment(null) or JBiclustGESetupManager.setupJBiclustGEMethodsEnvironment("/path_chosen_by_user"); -
Initializes the jbiclustge.properties file:
JBiclustGESetupManager.setupJBiclustGEProperties("Path to R library"); -
Reset the previous configuration of JBiclustGE:
JBiclustGESetupManager.resetPreviousConfiguration();
Biclustering Algorithms
All the biclustering algorithms extends to a main abstract class (jbiclustge.methods.algorithms.AbstractBiclusteringAlgorithmCaller), that contains all the methods that are necessary to establish a biclustering method in this API. Figure below shows a generic UML scheme concerning to all the biclustering methods integrated in this API.
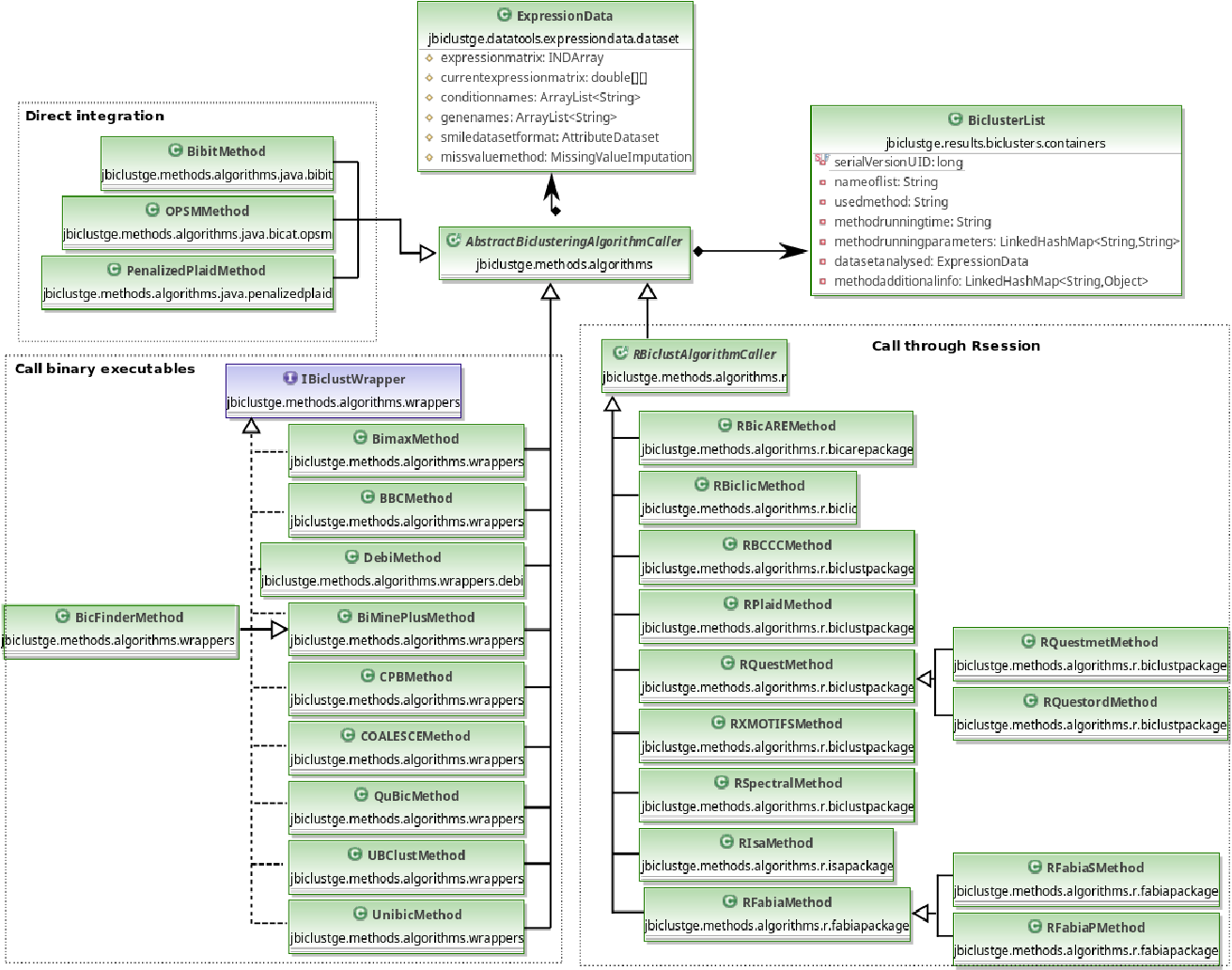
Extending JBiclustGE with new biclustering methods:
All the biclustering algorithms have to extend to the abstract class "AbstractBiclusteringAlgorithmCaller". However, JBiclustGE follows a given structure (presented in figure above) for the incorporation of new biclustering methods, which is:
- All the biclustering methods implemented in Java programming language extends directly to "AbstractBiclusteringAlgorithmCaller".
- All the biclustering methods implemented in R programming language extends to class "RBiclustAlgorithmCaller".
- All the biclustering methods implemented as binary executables using C, C++, Java, or other programming languages, have to extend to "AbstractBiclusteringAlgorithmCaller" and implement the interface "IBiclustWrapper".
Note: in order to avoid the definition of environment variables in Java to external libraries (e.g. libraries produced by Java Native Interface), is advisable to create a parsing routine that transforms the output (file with the resulting biclusters) of a given algorithm to the format used by the JBiclustGE.
If you want to extend the JBiclustGE-GUI by including your new biclustering method, follow this instructions
General procedure to add a new biclustering method to JBiclustGE
Figure below presents the main steps to integrate a new biclustering method in JBiclustGE API.

Abstract class AbstractBiclusteringAlgorithmCaller
Next figure shows the main methods that are imposed to the subclasses of AbstractBiclusteringAlgorithmCaller. Thus, when the new biclustering method class is extended to this abstract class, these methods should be adapted properly to the respective biclustering method.

Important methods that must be take into consideration when a class is extended to AbstractBiclusteringAlgorithmCaller:
-
The implemented class must be able to read a configuration of a given biclustering algorithm from a text file, which is loaded through method "setAlgorithmProperties(Properties props)". The configurations in that text file are set by a key=value parameterization, as presented below:
algorithm_parameter_john_doe1 = something_1 algorithm_parameter_john_doe2 = something_2 -
The implemented class must allow to instantiate an object "AlgorithmProperties" containing the configurations of biclustering algorithm. Because is that object that will allow to write these configurations to a text file, or use them in internal operations of JBiclustGE. Adjust method "AlgorithmProperties getAlgorithmAllowedProperties()" to your algorithm.
Abstract class RBiclustAlgorithmCaller
Biclustering methods developed in R programming language can be used by creating the respective java class that extends to RBiclustAlgorithmCaller. This abstract class includes several methods to link the Java environment to R environment through Rsession library. Thus, its necessary to have some knowledge about this library, such as, how the R commands can be defined in Rsession and how the objects produced in R environment can be returned to Java environment or vice versa. For that visit the Rsession website that contains all the instructions to perform such operations.
Figure presented below shows the main methods imposed by this class to their subclasses. There are two methods that need to be highlighted:
ArrayList<RPackageInfo> requiredLibraries() <-- defines which R packages are needed to load in R environment, in order to execute the biclustering algorithm.
ArrayList<String> loadSources() <-- loads a given R source file in R environment, which can be useful to load/run several functions by means of a R script, or even to load a biclustering algorithm implemented in a R source file.
It is also important to emphasize that it is necessary to create a method that can return the biclustering results from R environment, and convert them into a BiclusterList object. Such operations must be created using the Rsession library.

Interface IBiclustWrapper
This interface forces to assign the name of the executable of the biclustering method and the path for such binary executable.
BiclusterList and BiclusterResult objects
The biclustering results must be transformed to a "BiclusterList" object that agregates each one of the biclusters separately, in a object denominated as "BiclusterResult". Thus, each implemented algorithm has to build a "BiclusterList" object that aggregates the results of such algorithm, following a procedure similar to the one shown in the figure below.

How to run a biclustering method:
BicatYeast.csv dataset for example presented bellow, download
public static void main(String[] args) throws FileNotFoundException, IOException, MissingValueImputationException, ParseException {
URL dataUrl=HowToRunBiclusteringMethods.class.getResource("/BicatYeast.csv");
// load expression dataset from file //
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
// Example how to run CPB algorithm //
CPBMethod cpb=new CPBMethod(expressiondataset);
// for instance change number cluster to seed//
cpb.setNumberBiclustersToSeed(200);
// run algorithm //
cpb.run();
// get results obtained by the algorithm //
BiclusterList cpblistresults=cpb.getBiclusterResultList();
//print the results//
cpblistresults.printResults();
///////////////////////// Run Plaid example ////////////////////////////////////////
// methods that were developed in R or belongs to R package starts with R letter //
// initialize plaid method //
RPlaidMethod plaid=new RPlaidMethod();
// add expression dataset //
plaid.setExpressionData(expressiondataset);
// change the parameters that needed to be changed... //
plaid.addClusterType(BCPlaidClusterType.COLUMNS);
plaid.run();
// Use this function after running a algorithm that uses R environment, this allows to shutdown the R session. //
RConnector.closeSession();
BiclusterList plaidresults=plaid.getBiclusterResultList();
}
How to run the biclustering methods in MultiThread
public static void main(String[] args) throws Exception {
URL dataUrl=HowToRunBiclusteringMethods.class.getResource("/BicatYeast.csv");
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
/*
* Configure methods
*/
/*
* initialize list of task with methods that will be run in parallel
*/
ArrayList<BiclusteringTask> methodstasklist=new ArrayList<>();
/*
* add each task one by one
*/
BibitMethod bibit=new BibitMethod(expressiondataset);
methodstasklist.add(new BiclusteringTask(bibit));
RFabiaMethod fabia=new RFabiaMethod(expressiondataset);
methodstasklist.add(new BiclusteringTask(fabia));
/*
* Or use BiclusteringTasksFunctions to create a list of tasks
*/
RIsaMethod isa=new RIsaMethod(expressiondataset);
CPBMethod cpb=new CPBMethod(expressiondataset);
QuBicMethod quibic=new QuBicMethod(expressiondataset);
UnibicMethod unibic=new UnibicMethod(expressiondataset);
methodstasklist.addAll(BiclusteringTasksFunctions.createListOfBiclusteringTasks(isa,cpb,quibic,unibic));
/*
* After creating the list of tasks, use the MultiThreadBiclusteringTaskExecutor to execute these tasks
*/
// the output is the list of biclusters provided by the diferent biclustering algorithms, in the same order of the list of tasks.
List<BiclusterList> results=MultiThreadBiclusteringTaskExecutor.run(4, methodstasklist);
for (int i = 0; i < results.size(); i++) {
results.get(i).printResults();
}
/*
* Close R session connection
*/
RConnector.closeSession();
}
Executing the Gene Set Enrichment analysis engines
ecoli_example.csv dataset for example presented bellow, download
Ontologizer
public static void main(String[] args) throws Exception {
URL dataUrl=OntologizerExample.class.getResource("/ecoli_example.csv");
// load expression dataset from file
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
RIsaMethod isa=new RIsaMethod(expressiondataset);
isa.run();
/*
* initialize ontologizer analyser
*/
OntologizerEnrichmentAnalyser analyser=new OntologizerEnrichmentAnalyser(isa.getBiclusterResultList(), StandardAnnotationFile.Escherichiacoli);
/*
* setup ontologizer parameters
*/
analyser.addCalculationMethod(OntologizerCalculationMethod.TermForTerm);
analyser.addMultipleTestcorrectionMethod(OntologizerMTCMethod.Bonferroni);
analyser.run();
/*
* get results processed by ontologizer
*/
EnrichmentAnalysisResultList results=analyser.getEnrichmentAnalysisResults();
/*
* filter results with a pvalue lower than 0.01 and with adjusted pvalues
*/
results.filterAndProcessResults(0.01, true);
/*
* get only biclusters that have enriched genes
*/
EnrichedBiclusterList enrichedbiclusters= results.getEnrichedBiclusterList();
enrichedbiclusters.printResults();
}
topGO
public static void main(String[] args) throws Exception {
URL dataUrl=HowToRunBiclusteringMethods.class.getResource("/ecoli_example.csv");
// load expression dataset from file
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
RIsaMethod isa=new RIsaMethod(expressiondataset);
isa.run();
TopGOEnrichmentAnalyser analyser=new TopGOEnrichmentAnalyser(isa.getBiclusterResultList(), "org.EcK12.eg.db", true, TopGOMappingType.Symbol);
analyser.run();
EnrichmentAnalysisResultList results=analyser.getEnrichmentAnalysisResults();
/*
* process results with a pvalue lower than 0.05
*/
results.filterAndProcessResults(0.05, false);
System.out.println(results.getPercentageEnrichedBiclusters());
}
Post-processing Analysis
Coherence
public static void main(String[] args) throws FileNotFoundException, IOException, MissingValueImputationException, ParseException {
URL dataUrl=OntologizerExample.class.getResource("/ecoli_example.csv");
// load expression dataset from file
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
// run unibic method over dataset
UnibicMethod unibic=new UnibicMethod(expressiondataset);
unibic.run();
/*
* Perform coherence anlaysis
*/
// check multiplicative variance in all biclusters //
LinkedHashMap<Integer, Double> multivar= CoherenceAnalyser.getMultiplicativeVarianceForBiclusterList(unibic.getBiclusterResultList(), AnalysisTypeDimension.BOTH);
for (Integer bic : multivar.keySet()) {
System.out.println("Multiplicative variance of bicluster "+(bic+1)+" its: "+multivar.get(bic));
}
System.out.println("\n\n");
// check additive variance in all biclusters //
LinkedHashMap<Integer, Double> addvar= CoherenceAnalyser.getAdditiveVarianceForBiclusterList(unibic.getBiclusterResultList(), AnalysisTypeDimension.BOTH);
for (Integer bic : addvar.keySet()) {
System.out.println("Additive variance of bicluster "+(bic+1)+" its: "+addvar.get(bic));
}
System.out.println("\n\n");
// check constant variance in all biclusters //
LinkedHashMap<Integer, Double> constvar= CoherenceAnalyser.getConstantVarianceForBiclusterList(unibic.getBiclusterResultList(), AnalysisTypeDimension.BOTH);
for (Integer bic : constvar.keySet()) {
System.out.println("Constant variance of bicluster "+(bic+1)+" its: "+constvar.get(bic));
}
System.out.println("\n\n");
// check sign variance in all biclusters //
LinkedHashMap<Integer, Double> signvar= CoherenceAnalyser.getSignVarianceForBiclusterList(unibic.getBiclusterResultList(), AnalysisTypeDimension.BOTH);
for (Integer bic : signvar.keySet()) {
System.out.println("Sign variance of bicluster "+(bic+1)+" its: "+signvar.get(bic));
}
}
Coverage
public static void main(String[] args) throws FileNotFoundException, IOException, MissingValueImputationException, ParseException {
URL dataUrl=OntologizerExample.class.getResource("/ecoli_example.csv");
// load expression dataset from file //
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
// run unibic method over dataset //
UnibicMethod unibic=new UnibicMethod(expressiondataset);
unibic.run();
CoverageAnalyser coverage=new CoverageAnalyser(unibic.getBiclusterResultList());
System.out.println("Total genes coverage: "+coverage.getTotalgenescoverage()*100+"%\n");
System.out.println("Total conditions coverage: "+coverage.getTotalconditionscoverage()*100+"%\n");
System.out.println("Total matrix coverage: "+coverage.getTotalmatrixcoverage()*100+"%\n");
}
Overlap
public static void main(String[] args) throws Exception {
// change log level //
LogMessageCenter.getLogger().setLogLevel(MTULogLevel.DEBUG);
URL dataUrl=OntologizerExample.class.getResource("/ecoli_example.csv");
// load expression dataset from file //
ExpressionData expressiondataset=ExpressionData.importFromTXTFormat(dataUrl.getFile()).load();
// run unibic method over dataset //
CPBMethod cpb=new CPBMethod(expressiondataset);
cpb.run();
BiclusterList results=cpb.getBiclusterResultList();
// check overlap between bicluster 1 and 2
System.out.println("overlap between bicluster 1 and 2: "+OverlapAnalyser.getOverlapBeetwenTwoBiclusters(results.get(0), results.get(1)));
// remove biclusters that have an overlap higher than a threshold
BiclusterList filteredlist=OverlapAnalyser.filterBiclusterListWithOverlapThreshold(results, 0.2, results.size());
filteredlist.printResults();
System.out.println("Average Overlap: "+OverlapAnalyser.AverageOverlap(results));
}